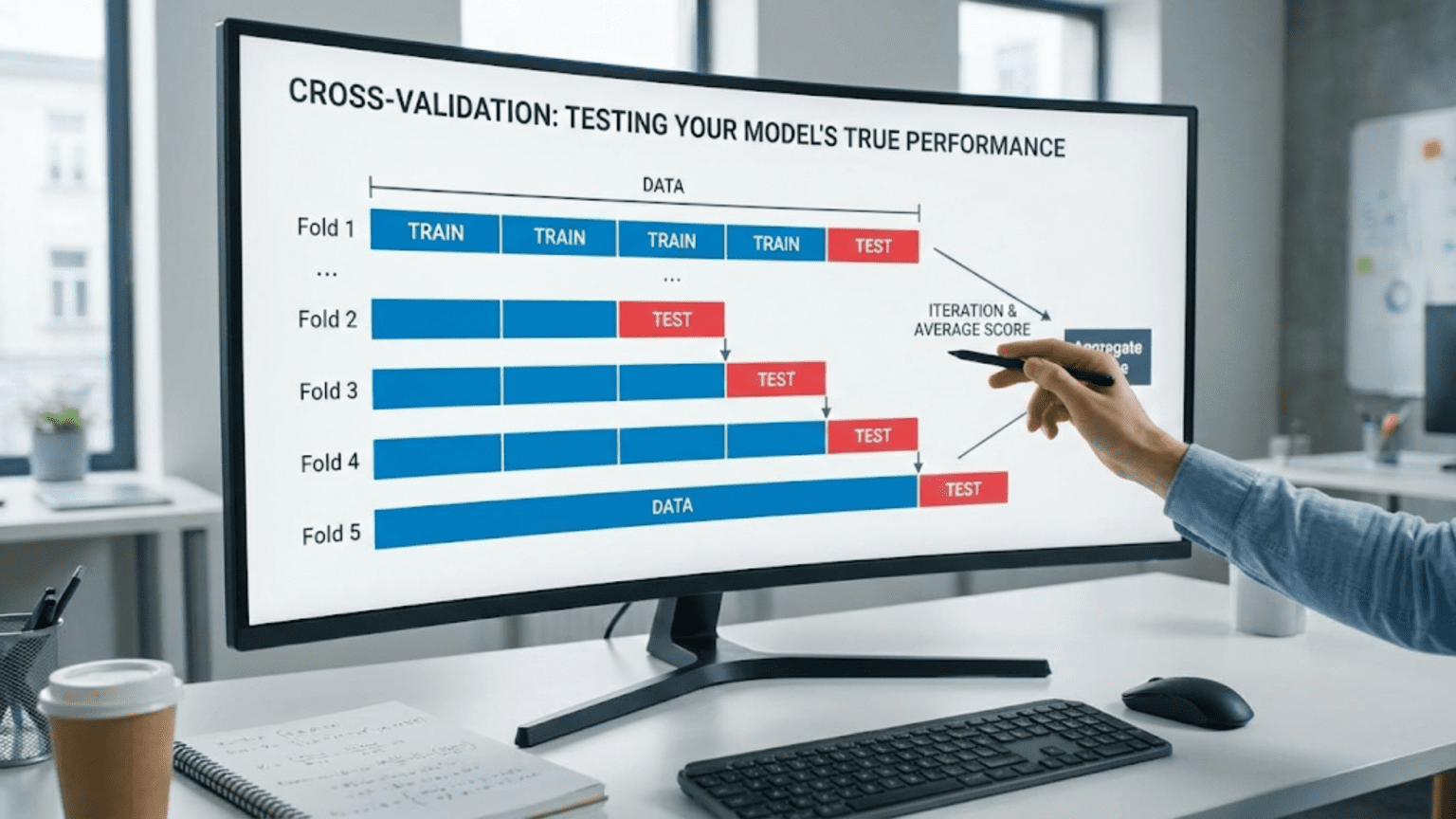
Cross-validation is a resampling technique that evaluates machine learning models by training and testing on multiple different data splits to obtain more reliable performance estimates. The most common method, k-fold cross-validation, divides data into k subsets (folds), trains on k-1 folds and tests on the remaining fold, repeating k times with different test folds. This produces k performance scores that are averaged for a robust estimate, revealing whether good performance on a single split was luck or genuine model quality.
Introduction: The Problem with Single Splits
Imagine evaluating a student’s knowledge by asking just one question. They might happen to know that specific topic well—or poorly—giving you a misleading impression of their overall understanding. You’d get a much more accurate assessment by asking multiple questions covering different topics and averaging the results.
Machine learning faces exactly this challenge. When you split data into a single train-test set and evaluate once, performance depends partly on luck—which specific examples ended up in which set. You might get fortunate with an easy test set, yielding overly optimistic estimates. Or unlucky with a particularly challenging test set, underestimating your model’s true capability.
This single-split evaluation has high variance. Train the same model on different random splits, and you’ll get different performance scores—sometimes dramatically different. A model might achieve 85% accuracy on one split, 78% on another, and 91% on a third. Which number represents true performance? You simply don’t know.
Cross-validation solves this problem by evaluating models on multiple different splits and averaging the results. Instead of one train-test split giving one performance estimate, you get multiple estimates from multiple splits, producing a more stable, reliable assessment of true model quality.
This comprehensive guide explores cross-validation in depth. You’ll learn why it matters, how different methods work, when to use which approach, how to implement them properly, and common pitfalls to avoid. Whether you’re comparing models, tuning hyperparameters, or seeking rigorous performance estimates, understanding cross-validation is essential for reliable machine learning.
Why Cross-Validation Matters
Cross-validation addresses several critical challenges in model evaluation.
Problem 1: Variance in Performance Estimates
Single Split Example:
Same model, different random splits:
Split 1: 82% accuracy
Split 2: 88% accuracy
Split 3: 79% accuracy
Split 4: 85% accuracy
Split 5: 91% accuracy
Which is the "true" performance?
Range: 79% to 91% (12 percentage points!)High Variance: Single split gives unreliable estimate
Cross-Validation Solution:
5-fold cross-validation:
Fold 1: 82%
Fold 2: 88%
Fold 3: 79%
Fold 4: 85%
Fold 5: 91%
Average: 85%
Standard deviation: 4.5%
Report: 85% ± 4.5%Lower Variance: Averaged estimate more reliable
Problem 2: Inefficient Data Use
Traditional Split:
1000 total examples
700 training (70%)
300 test (30%)
Only 70% of data used for training
Only 30% evaluatedCross-Validation:
5-fold CV uses all 1000 examples:
- Each example used for training in 4 folds
- Each example tested in 1 fold
- 100% data utilizationEspecially valuable with limited data.
Problem 3: Lucky/Unlucky Splits
Lucky Split:
By chance, easy examples in test set
Model achieves 95% accuracy
Overestimates true performance
Deploy model → actual performance is 80%
Disappointing!Unlucky Split:
By chance, hard examples in test set
Model achieves 75% accuracy
Underestimates true performance
Reject good model
Miss opportunity!Cross-Validation: Averages across multiple splits, reducing luck factor
Problem 4: Small Dataset Reliability
Challenge: With only 200 examples:
Traditional 70-30 split:
140 training
60 test
Test set too small for reliable estimate
High variance in test performanceCross-Validation: 10-fold CV:
Each fold: 180 training, 20 test
Average 10 folds → more stable estimate
Better use of limited dataK-Fold Cross-Validation: The Standard Approach
The most widely used cross-validation method.
How K-Fold Works
Process:
- Divide data into K equal folds (typically K=5 or K=10)
- For each fold k = 1 to K:
- Use fold k as test set
- Use remaining K-1 folds as training set
- Train model on training folds
- Evaluate on test fold
- Record performance score
- Average K performance scores
- Report mean and standard deviation
Visual Example (5-Fold):
Iteration 1: [Test][Train][Train][Train][Train] → Score: 82%
Iteration 2: [Train][Test][Train][Train][Train] → Score: 85%
Iteration 3: [Train][Train][Test][Train][Train] → Score: 88%
Iteration 4: [Train][Train][Train][Test][Train] → Score: 84%
Iteration 5: [Train][Train][Train][Train][Test] → Score: 86%
Mean: 85%
Std: 2.3%Implementation Example
Using Scikit-learn:
from sklearn.model_selection import cross_val_score
from sklearn.ensemble import RandomForestClassifier
# Initialize model
model = RandomForestClassifier(n_estimators=100)
# Perform 5-fold CV
scores = cross_val_score(model, X, y, cv=5, scoring='accuracy')
# Results
print(f"Scores: {scores}")
print(f"Mean: {scores.mean():.3f}")
print(f"Std: {scores.std():.3f}")
print(f"95% CI: {scores.mean():.3f} ± {1.96*scores.std():.3f}")Output:
Scores: [0.82, 0.85, 0.88, 0.84, 0.86]
Mean: 0.850
Std: 0.023
95% CI: 0.850 ± 0.045Choosing K
Common Values:
- K=5: Faster, good balance
- K=10: More thorough, standard choice
- K=n (Leave-One-Out): Maximum data use, computationally expensive
Tradeoffs:
Small K (K=3):
- Faster computation (fewer iterations)
- Less data for training each fold
- Higher variance in estimates
- Use when: Computational resources limited
Large K (K=10):
- More computation (more iterations)
- More data for training each fold
- Lower variance in estimates
- Use when: Want reliable estimates
Very Large K (K=20+):
- Expensive computation
- Diminishing returns
- Similar to K=10
- Rarely worth the cost
Recommendation: K=5 or K=10 for most applications
Interpreting Results
Mean Score: Central estimate of model performance
Standard Deviation: Variability across folds
- Small std: Consistent performance
- Large std: Performance varies by data split (concerning)
Example Interpretations:
Model A: 85% ± 2%
- Consistent, reliable
- Performance stable across splits
Model B: 85% ± 12%
- High variance
- Performance depends heavily on data split
- Concerning even with same meanStatistical Significance:
Model A: 85% ± 2%
Model B: 83% ± 2%
Difference: 2 percentage points
Are they significantly different?
Simple test: Check if confidence intervals overlap
A: [81%, 89%]
B: [79%, 87%]
Overlapping → Not clearly different
For rigorous testing: Paired t-test on fold scoresStratified K-Fold: Maintaining Class Distribution
For classification with imbalanced classes.
The Problem with Regular K-Fold
Imbalanced Dataset:
1000 examples: 900 class A, 100 class B (90%-10% split)
Regular 5-fold might create:
Fold 1: 95% A, 5% B
Fold 2: 88% A, 12% B
Fold 3: 92% A, 8% B
Fold 4: 87% A, 13% B
Fold 5: 88% A, 12% B
Each fold has different class distribution!
Unfair comparison, higher varianceStratified Solution
Stratified K-Fold: Each fold maintains overall class distribution
Example:
All folds have:
90% class A
10% class B
Fair comparison across folds
Lower variance in estimatesImplementation:
from sklearn.model_selection import StratifiedKFold
skf = StratifiedKFold(n_splits=5, shuffle=True, random_state=42)
for fold, (train_idx, test_idx) in enumerate(skf.split(X, y)):
X_train, X_test = X[train_idx], X[test_idx]
y_train, y_test = y[train_idx], y[test_idx]
# Check stratification
print(f"Fold {fold+1} - Train class distribution: {np.bincount(y_train)}")
print(f"Fold {fold+1} - Test class distribution: {np.bincount(y_test)}")When to Use:
- Classification problems
- Imbalanced classes
- Want consistent class distribution across folds
Default Choice: Use stratified k-fold for classification unless you have a reason not to
Time Series Cross-Validation: Respecting Temporal Order
For sequential, time-dependent data.
Why Regular CV Fails for Time Series
Problem: Regular k-fold randomly assigns examples to folds
Time Series Issue:
Training on future data to predict past = data leakage!
Example (wrong):
Jan-Mar data in test fold
Apr-Jun data in training folds
Using future to predict past → unrealistic, inflated performanceReal World: Always predict future from past, never vice versa
Time Series Split Approaches
Forward Chaining (Expanding Window)
Method: Progressively expand training set, always test on future
Structure:
Fold 1: Train[1-3], Test[4]
Fold 2: Train[1-4], Test[5]
Fold 3: Train[1-5], Test[6]
Fold 4: Train[1-6], Test[7]
Fold 5: Train[1-7], Test[8]
Training set grows, test always in futureImplementation:
from sklearn.model_selection import TimeSeriesSplit
tscv = TimeSeriesSplit(n_splits=5)
for fold, (train_idx, test_idx) in enumerate(tscv.split(X)):
print(f"Fold {fold+1}")
print(f" Train: indices {train_idx[0]} to {train_idx[-1]}")
print(f" Test: indices {test_idx[0]} to {test_idx[-1]}")When to Use:
- Training data quantity affects performance
- Want to simulate progressive learning
- Standard approach for time series
Sliding Window
Method: Fixed-size training window slides forward
Structure:
Fold 1: Train[1-3], Test[4]
Fold 2: Train[2-4], Test[5]
Fold 3: Train[3-5], Test[6]
Fold 4: Train[4-6], Test[7]
Fold 5: Train[5-7], Test[8]
Training window size constantWhen to Use:
- Recent data more relevant (concept drift)
- Want to evaluate with fixed training size
- Older data becomes less relevant
Implementation Example
Stock Price Prediction:
import pandas as pd
from sklearn.model_selection import TimeSeriesSplit
# Time series data
data = pd.DataFrame({
'date': pd.date_range('2020-01-01', periods=1000, freq='D'),
'price': stock_prices,
'features': feature_data
})
# Sort by time (critical!)
data = data.sort_values('date')
# Time series CV
tscv = TimeSeriesSplit(n_splits=5)
scores = []
for train_idx, test_idx in tscv.split(data):
train = data.iloc[train_idx]
test = data.iloc[test_idx]
# Train model
model.fit(train[features], train['price'])
# Evaluate on future data
score = model.score(test[features], test['price'])
scores.append(score)
print(f"Train: {train['date'].min()} to {train['date'].max()}")
print(f"Test: {test['date'].min()} to {test['date'].max()}")
print(f"Score: {score:.3f}\n")
print(f"Mean Score: {np.mean(scores):.3f}")Leave-One-Out Cross-Validation (LOOCV)
Extreme case where K = n (number of examples).
How LOOCV Works
Process:
- Train on n-1 examples
- Test on 1 example
- Repeat n times (each example as test once)
- Average n scores
Example (10 examples):
Iteration 1: Train[2-10], Test[1]
Iteration 2: Train[1,3-10], Test[2]
Iteration 3: Train[1-2,4-10], Test[3]
...
Iteration 10: Train[1-9], Test[10]Advantages
- Maximum training data: Uses n-1 examples for training
- No randomness: Deterministic (no shuffle needed)
- Unbiased estimate: Each example tested exactly once
Disadvantages
- Computationally expensive: n training runs for n examples
- High variance: Each test set has only 1 example
- Not practical for large datasets
Example Computation:
Dataset with 10,000 examples
LOOCV requires 10,000 training runs!
Compare to:
5-fold CV: 5 training runs
10-fold CV: 10 training runsWhen to Use LOOCV
Good For:
- Very small datasets (n < 100)
- Maximum data efficiency critical
- Computationally cheap models (fast training)
Avoid When:
- Large datasets (too expensive)
- Slow-to-train models
- Need low-variance estimates
Implementation:
from sklearn.model_selection import LeaveOneOut
loo = LeaveOneOut()
scores = []
for train_idx, test_idx in loo.split(X):
model.fit(X[train_idx], y[train_idx])
score = model.score(X[test_idx], y[test_idx])
scores.append(score)
print(f"Mean: {np.mean(scores):.3f}")
print(f"Std: {np.std(scores):.3f}")Nested Cross-Validation: Unbiased Hyperparameter Tuning
For rigorous evaluation when tuning hyperparameters.
The Problem with Standard CV + Hyperparameter Tuning
Typical (Biased) Approach:
1. Use cross-validation to tune hyperparameters
2. Report the best CV score as model performance
Problem: You've "learned" from the CV folds during tuning
The CV score is optimistically biased
True performance on new data will be lowerExample:
Try 100 hyperparameter combinations
Best achieves 87% on 5-fold CV
But: You selected this based on CV performance
Expected performance on truly new data: ~83%
You've overfit to the CV foldsNested CV Solution
Two Loops:
Outer Loop: Generates unbiased performance estimate
- Standard k-fold CV (e.g., 5 folds)
- Each fold treated as “test set”
Inner Loop: Performs hyperparameter tuning
- Nested k-fold CV on training portion (e.g., 3 folds)
- Selects best hyperparameters for that outer fold
Structure:
Outer Fold 1:
Train data: Outer folds 2-5
Inner CV: Tune hyperparameters on folds 2-5
Select best parameters
Test data: Outer fold 1
Evaluate with best parameters → Score 1
Outer Fold 2:
Train data: Outer folds 1, 3-5
Inner CV: Tune hyperparameters
Select best parameters
Test data: Outer fold 2
Evaluate → Score 2
...
Outer Fold 5:
...
Evaluate → Score 5
Final estimate: Average of 5 outer scoresImplementation
Manual Implementation:
from sklearn.model_selection import cross_val_score, GridSearchCV
from sklearn.ensemble import RandomForestClassifier
# Outer CV
outer_cv = StratifiedKFold(n_splits=5, shuffle=True, random_state=42)
outer_scores = []
for train_idx, test_idx in outer_cv.split(X, y):
X_train_outer, X_test_outer = X[train_idx], X[test_idx]
y_train_outer, y_test_outer = y[train_idx], y[test_idx]
# Inner CV for hyperparameter tuning
inner_cv = StratifiedKFold(n_splits=3, shuffle=True, random_state=42)
param_grid = {
'n_estimators': [50, 100, 200],
'max_depth': [5, 10, 15]
}
grid_search = GridSearchCV(
RandomForestClassifier(),
param_grid,
cv=inner_cv,
scoring='accuracy'
)
# Tune on outer training fold
grid_search.fit(X_train_outer, y_train_outer)
# Evaluate best model on outer test fold
score = grid_search.score(X_test_outer, y_test_outer)
outer_scores.append(score)
print(f"Best params: {grid_search.best_params_}")
print(f"Outer fold score: {score:.3f}\n")
print(f"Nested CV Score: {np.mean(outer_scores):.3f} ± {np.std(outer_scores):.3f}")Using cross_validate:
from sklearn.model_selection import cross_validate
# GridSearchCV acts as the estimator
grid_search = GridSearchCV(
RandomForestClassifier(),
param_grid={'n_estimators': [50, 100, 200], 'max_depth': [5, 10, 15]},
cv=3 # Inner CV
)
# Outer CV
nested_scores = cross_val_score(
grid_search,
X, y,
cv=5, # Outer CV
scoring='accuracy'
)
print(f"Nested CV: {nested_scores.mean():.3f} ± {nested_scores.std():.3f}")When to Use Nested CV
Essential For:
- Rigorous performance estimates
- Academic publications
- Critical applications (medical, financial)
- When hyperparameter tuning is involved
- Comparing fundamentally different approaches
Overkill For:
- Quick prototyping
- Computationally expensive (nested CV takes 25-50x longer)
- Clear winners without rigorous testing
Tradeoff: Unbiased estimates vs. computational cost
Group K-Fold: Keeping Related Examples Together
For grouped data where examples aren’t independent.
The Problem
Scenario: Medical data with multiple scans per patient
Regular K-Fold Problem:
Patient A has 10 scans
Random k-fold might put:
- 7 scans in training
- 3 scans in test
Model learns Patient A's specific characteristics
Inflated performance (recognizing same patient)
Doesn't reflect real-world (new patients)Group K-Fold Solution
Ensure all examples from same group in same fold
Example:
Patients: A, B, C, D, E (each with multiple scans)
Fold 1: Train[B,C,D,E], Test[A] (all scans from A in test)
Fold 2: Train[A,C,D,E], Test[B]
Fold 3: Train[A,B,D,E], Test[C]
Fold 4: Train[A,B,C,E], Test[D]
Fold 5: Train[A,B,C,D], Test[E]
No patient in both training and testImplementation:
from sklearn.model_selection import GroupKFold
# Groups indicate which patient each scan belongs to
groups = [1, 1, 1, 2, 2, 3, 3, 3, 3, 4, 4, 5, 5, 5]
# Patient 1: scans 0-2
# Patient 2: scans 3-4
# etc.
gkf = GroupKFold(n_splits=5)
for train_idx, test_idx in gkf.split(X, y, groups=groups):
print(f"Train groups: {set(groups[i] for i in train_idx)}")
print(f"Test groups: {set(groups[i] for i in test_idx)}")When to Use
Required For:
- Multiple measurements per patient/customer/sensor
- Time series data for multiple entities
- Hierarchical data (students within schools)
- Any non-independent observations
Examples:
- Medical: Multiple scans/tests per patient
- Finance: Multiple transactions per customer
- IoT: Multiple readings per sensor
- Social: Multiple posts per user
Practical Example: Complete Cross-Validation Workflow
Let’s walk through a complete example.
Problem Setup
Task: Predict customer churn Data: 5,000 customers Features: 25 features (demographics, usage, billing) Label: Binary (churned=1, retained=0) Class Distribution: 20% churned, 80% retained (imbalanced)
Step 1: Initial Evaluation with Simple Split
Baseline:
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestClassifier
# Simple 80-20 split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=42
)
model = RandomForestClassifier()
model.fit(X_train, y_train)
score = model.score(X_test, y_test)
print(f"Test Accuracy: {score:.3f}")Result: Test Accuracy: 0.847
Problem: Single split, could be lucky/unlucky
Step 2: 5-Fold Cross-Validation
Better Estimate:
from sklearn.model_selection import cross_val_score
scores = cross_val_score(
RandomForestClassifier(),
X, y,
cv=5,
scoring='accuracy'
)
print(f"CV Scores: {scores}")
print(f"Mean: {scores.mean():.3f} ± {scores.std():.3f}")Result:
CV Scores: [0.832, 0.858, 0.841, 0.829, 0.855]
Mean: 0.843 ± 0.012Insight: Average 0.843, consistent across folds (low std)
Step 3: Stratified K-Fold (Account for Imbalance)
Maintain Class Distribution:
from sklearn.model_selection import StratifiedKFold
skf = StratifiedKFold(n_splits=5, shuffle=True, random_state=42)
scores = cross_val_score(
RandomForestClassifier(),
X, y,
cv=skf,
scoring='accuracy'
)
print(f"Stratified CV: {scores.mean():.3f} ± {scores.std():.3f}")Result: Stratified CV: 0.844 ± 0.011
Comparison:
Regular CV: 0.843 ± 0.012
Stratified CV: 0.844 ± 0.011
Similar but stratified slightly more stableStep 4: Multiple Metrics
Don’t Rely on Accuracy Alone:
from sklearn.model_selection import cross_validate
scoring = {
'accuracy': 'accuracy',
'precision': 'precision',
'recall': 'recall',
'f1': 'f1',
'roc_auc': 'roc_auc'
}
results = cross_validate(
RandomForestClassifier(),
X, y,
cv=StratifiedKFold(n_splits=5, shuffle=True, random_state=42),
scoring=scoring,
return_train_score=True
)
for metric in scoring.keys():
test_scores = results[f'test_{metric}']
print(f"{metric}: {test_scores.mean():.3f} ± {test_scores.std():.3f}")Results:
accuracy: 0.844 ± 0.011
precision: 0.791 ± 0.023
recall: 0.712 ± 0.031
f1: 0.749 ± 0.021
roc_auc: 0.892 ± 0.015Insight:
- Good accuracy and AUC
- Moderate precision and recall
- Recall lower (missing some churners)
Step 5: Hyperparameter Tuning with Nested CV
Rigorous Evaluation:
from sklearn.model_selection import GridSearchCV
# Hyperparameter grid
param_grid = {
'n_estimators': [50, 100, 200],
'max_depth': [5, 10, 15, None],
'min_samples_split': [2, 5, 10]
}
# Inner CV for tuning
inner_cv = StratifiedKFold(n_splits=3, shuffle=True, random_state=42)
grid_search = GridSearchCV(
RandomForestClassifier(),
param_grid,
cv=inner_cv,
scoring='f1'
)
# Outer CV for unbiased estimate
outer_cv = StratifiedKFold(n_splits=5, shuffle=True, random_state=42)
nested_scores = cross_val_score(
grid_search,
X, y,
cv=outer_cv,
scoring='f1'
)
print(f"Nested CV F1: {nested_scores.mean():.3f} ± {nested_scores.std():.3f}")Results:
Nested CV F1: 0.756 ± 0.019Comparison:
Non-nested (biased): 0.749 ± 0.021
Nested CV (unbiased): 0.756 ± 0.019
Nested actually slightly better here
(Sometimes nested is lower due to removing optimistic bias)Step 6: Final Model Training
After CV confirms good performance:
# Train on all available data
grid_search.fit(X, y)
print(f"Best parameters: {grid_search.best_params_}")
print(f"Best CV score: {grid_search.best_score_:.3f}")
# Final model ready for deployment
final_model = grid_search.best_estimator_Results:
Best parameters: {
'n_estimators': 200,
'max_depth': 15,
'min_samples_split': 5
}
Best CV score: 0.758
Expected production performance: ~0.756 (from nested CV)Common Pitfalls and Best Practices
Pitfall 1: Data Leakage in Preprocessing
Wrong:
# Normalize entire dataset
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
X_scaled = scaler.fit_transform(X) # Uses all data!
# Then cross-validate
scores = cross_val_score(model, X_scaled, y, cv=5)
# Biased! Test folds influenced by their own dataRight:
from sklearn.pipeline import Pipeline
# Pipeline ensures preprocessing happens within CV
pipeline = Pipeline([
('scaler', StandardScaler()),
('model', RandomForestClassifier())
])
scores = cross_val_score(pipeline, X, y, cv=5)
# Correct! Each fold preprocessed independentlyPitfall 2: Not Shuffling Data
Problem: Data might be ordered
Example:
First 80% examples: Class A
Last 20% examples: Class B
Without shuffling:
Some folds have only Class A
Some folds have only Class B
Invalid evaluationSolution: Always shuffle (except time series)
kf = KFold(n_splits=5, shuffle=True, random_state=42)Pitfall 3: Different Random States
Problem: Comparing models with different random splits
Wrong:
# Model A
scores_A = cross_val_score(modelA, X, y, cv=5) # Random state varies
# Model B (different splits!)
scores_B = cross_val_score(modelB, X, y, cv=5)
# Unfair comparisonRight:
# Use same CV splitter for both
cv = StratifiedKFold(n_splits=5, shuffle=True, random_state=42)
scores_A = cross_val_score(modelA, X, y, cv=cv)
scores_B = cross_val_score(modelB, X, y, cv=cv)
# Fair comparison, same foldsPitfall 4: Using Test Set Multiple Times
Problem: Evaluating and tuning based on test performance
Wrong:
# Try different models, check test performance each time
for model in models:
test_score = evaluate_on_test_set(model)
if test_score > best_score:
best_model = model
# You've overfit to test set through selectionRight:
# Use CV for model selection
for model in models:
cv_score = cross_val_score(model, X_train, y_train, cv=5).mean()
if cv_score > best_score:
best_model = model
# Only evaluate best model on test set once
final_score = best_model.score(X_test, y_test)Pitfall 5: Ignoring Computational Cost
Problem: Nested CV with expensive models
Example:
Nested CV: 5 outer × 3 inner = 15 folds
Grid search: 100 parameter combinations
Total training runs: 15 × 100 = 1,500
If each takes 10 minutes: 250 hours!Solutions:
- Use smaller inner CV (2-3 folds)
- Random search instead of grid search
- Reduce parameter grid
- Use simpler models
- Sample data for hyperparameter tuning
Pitfall 6: Wrong CV for Time Series
Problem: Using regular k-fold for temporal data
Example:
Stock price prediction with regular k-fold
→ Training on future data
→ Inflated performance estimates
→ Fails in productionSolution: Always use TimeSeriesSplit for temporal data
Advanced Cross-Validation Techniques
Repeated K-Fold
Idea: Run k-fold multiple times with different random states
Process:
5-fold CV repeated 3 times = 15 evaluations
Average all 15 scores
Even more stable estimateImplementation:
from sklearn.model_selection import RepeatedStratifiedKFold
rskf = RepeatedStratifiedKFold(n_splits=5, n_repeats=3, random_state=42)
scores = cross_val_score(model, X, y, cv=rskf)
print(f"15 scores from 3 repeats of 5-fold CV")
print(f"Mean: {scores.mean():.3f} ± {scores.std():.3f}")When to Use:
- Small datasets (need maximum reliability)
- High-stakes decisions
- Small performance differences to detect
Monte Carlo CV (Shuffle-Split)
Idea: Random train-test splits repeated many times
Difference from K-Fold: Splits are random, not exhaustive
Implementation:
from sklearn.model_selection import ShuffleSplit
ss = ShuffleSplit(n_splits=10, test_size=0.2, random_state=42)
scores = cross_val_score(model, X, y, cv=ss)Advantages:
- Control train/test sizes independently
- Can repeat many times
- More randomness in estimates
Disadvantages:
- Some examples may never be in test set
- Some examples may be in test set multiple times
- Less efficient than k-fold
Comparison: Cross-Validation Methods
| Method | Best For | Pros | Cons | Computation |
|---|---|---|---|---|
| K-Fold | General purpose | Balanced, efficient | Variance in small K | K × training |
| Stratified K-Fold | Imbalanced classification | Maintains distribution | Only for classification | K × training |
| Time Series CV | Temporal data | Respects time order | Can’t shuffle | K × training |
| LOOCV | Small datasets | Maximum data use | High variance, expensive | n × training |
| Group K-Fold | Grouped data | Prevents leakage | Reduced effective samples | K × training |
| Nested CV | Rigorous evaluation + tuning | Unbiased estimates | Very expensive | (K_outer × K_inner × grid_size) × training |
| Repeated K-Fold | Maximum reliability | Most stable estimates | Expensive | (K × repeats) × training |
Best Practices Summary
Always Do
- Use cross-validation for model selection
- Shuffle data (except time series)
- Set random_state for reproducibility
- Use stratified CV for classification
- Use pipelines to prevent data leakage
- Report mean and standard deviation
- Use same CV splits when comparing models
Consider
- Nested CV for rigorous hyperparameter tuning
- Multiple metrics beyond single score
- Repeated CV for critical decisions
- Appropriate CV for data structure (groups, time series)
- Confidence intervals for statistical rigor
Never Do
- Preprocess before splitting (causes leakage)
- Use test set during development
- Compare models on different splits
- Ignore standard deviation (just reporting mean)
- Use regular CV for time series
Conclusion: The Foundation of Reliable Evaluation
Cross-validation transforms model evaluation from a single, potentially misleading number into a robust, reliable estimate of true performance. By training and testing on multiple different data splits, you average out the luck factor inherent in any single split, revealing whether good performance is genuine or just fortunate data sampling.
The key insights about cross-validation:
K-fold cross-validation is the standard approach, providing good balance between computational cost and estimate reliability. Use K=5 or K=10 for most applications.
Stratified k-fold maintains class distributions across folds, essential for imbalanced classification problems.
Time series CV respects temporal ordering, critical for sequential data where using future to predict past would leak information.
Nested cross-validation provides unbiased estimates when hyperparameter tuning is involved, separating model selection from evaluation.
Group k-fold keeps related examples together, preventing data leakage when observations aren’t independent.
Cross-validation isn’t just about getting better numbers—it’s about honest evaluation. A single train-test split might yield 90% accuracy through luck, leading you to deploy a model that actually performs at 80%. Cross-validation reveals the truth, averaging across multiple scenarios to estimate real-world performance.
The computational cost is worth it. Training 5 or 10 times instead of once seems expensive, but it’s far cheaper than deploying an overfit model, discovering it fails in production, and having to rebuild. Cross-validation catches problems during development, when fixing them is easy.
As you build machine learning systems, make cross-validation standard practice. Use it for model selection, hyperparameter tuning, and performance reporting. Report both mean and standard deviation to convey reliability. Use appropriate variants for your data structure. Prevent data leakage through proper pipelines.
Master cross-validation, and you’ve mastered the art of honest model evaluation—building systems whose development performance actually matches production reality, delivering the value you expect rather than disappointing with inflated estimates that never materialize in the real world.